Archive for the ‘molecules’ tag
Molecule Rigging
Some of you reading are familiar with using the epmv plugin within Cinema 4D. Gary Welsh came up with a workflow for rigging those downloaded molecules to make them dynamic. I haven’t tried this one myself yet, but it looks really cool. I wanted the reference for myself, so I thought that I would share it with anyone else reading as well.
If you go to the YouTube page directly, you can even download the .c4d file directly.
DNA Replication – 3D Animation
Hello again readers! I’m so happy to be able to finally share with you all an animation that I worked on some months back for Sapling Learning. Some of you will recall me writing about this one before.
The 3D models were created by studying the molecular sizes and shapes of the relevant molecules and then creating simplified representations. The 3D animation itself was created using Maxon’s Cinema 4D. Then the individual shots were composited together in Adobe’s After Effects. Audacity was used to record the narration, and then it was all put together for timing in Final Cut Pro (I still use an older version and cannot speak to all the changes they made in X.) I went back into After Effects for further labeling and effects (like the zoom into DNA polymerase) after that. So that’s the basic work flow.
There are a lot of biochemical processes that feel like a very complicated ballet as you get further into them. Putting together the action of everything happening at the same time for the final shot in this animation really got me thinking about that analogy again. And the truth is, that when this actually happens in nature, that lagging strand is actually being whipped around by helicase and read in an even more complicated fashion. I really like that we emphasized that we were simplifying for clarity with this one. A lot of educational materials would just let students find that out later if they went on to the next step in their studies. That kind of thing always bothered me when I learned about such processes in school, and I’m happy to be with content experts who are willing to take the time to either make something more accurate, or let students know when things are being brushed over or left out of the picture for clarity. I think that kind of thing is important.
Anyway, I’m really happy to be able to share this with you all.
Translation – The narrated version is up and public
Last week I got permission at Sapling to post the new animation. I went with the narrated version for now. I may post the silent one at a later date.
I learned a lot putting this together. I learned a good bit about working with cloners in Cinema 4D, and got to practice with C4D’s shaders, and even touched on coding a bit with trying to get that mRNA strand to move the way I wanted it to.
All in all, I think it came out really well, both versions actually. I’m proud of it. And I’m really enjoying working with such amazing and knowledgeable folks over at Sapling. I learn the coolest stuff from them, all the time.
Animation in Layers
Goodness me, it’s the 4th of July and I intend to go jump in a pool and play in a little while. But first I wanted to pop in and say a few words over here. Lately over at Sapling I’ve been pretty consumed with an animation that I’m putting together over there. It’s been a whole lot of work, and it’s nearly finished now. I get excited at times like these.
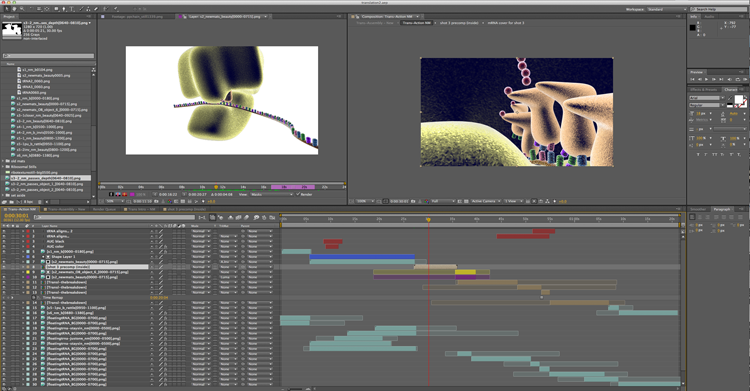
The animation is about the translation process. From a technical standpoint, it’s been interesting how much time I’ve been spending in the program, After Effects. I usually do a little compositing there, but then have generally brought my work into Final Cut Pro or something like that to finish up the actual shot arrangement. With this one, I’ve found it made more sense to actually just put the whole thing together in After Effects. I’m having to be really careful about organization and use a lot of pre-compositions. And even so, I don’t believe that I have ever taken on this many layers of video to wrangle for another project in my entire life. That’s the thing about animation though I guess. You literally make all of it from scratch.
One thing that I don’t care for in After Effects as opposed to your more editing and less effect driven software, is it’s insistence that every little thing have it’s own layer. This has been one of those pieces for which I’ve wanted a lot of layered subtlety going on behind the scene. It would have been nice to be able to dedicate a couple of tracks for that, rather than adding a new one for every piece of footage used. That being said though, After Effects does a great job at handling image sequence footage and you do have a great deal of control over a lot of details. Still, if anyone from Adobe happens to read this, maybe the next version could at least give us the ability to group tracks into folders. For when you don’t quite want to create a pre-composition and send your tracks to another timeline, but you still want to clean them up a bit for viewing sake, or to add some sort of matting to the whole bundle, just that one little thing would make After Effects so much better in general.
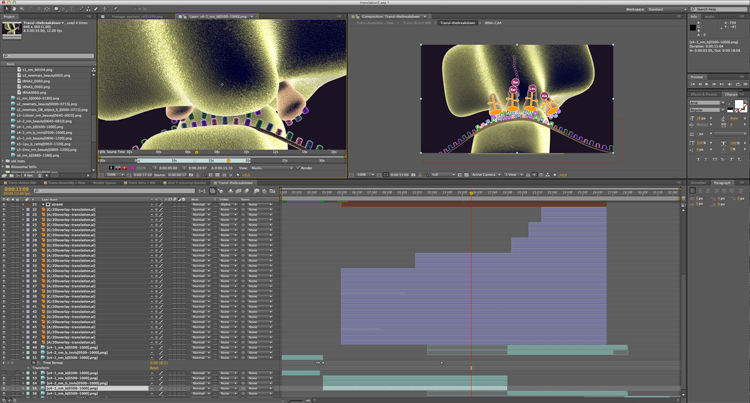
I especially thought that when I got into the part of this animation that I like to call the breakdown. Textbooks often use similar static illustrations to represent things that may or may not atomically look like the molecule in question. And really it’s only rather recently with the work of Dr. Venki Ramakrishnan (video link to a lecture of his about the ribosome and translation process) that we even fully know the exact structure of the ribosome. So one of the things that I’ve been excited to do with this one is to create some more standard looking images and then layer them over the 3D animation. I then keyframed the individual components to follow their 3D counterparts so we can actually see the connection between those representations and a more 3D space. I have been simultaneously irritated with myself for coming up with such a time consuming and intricate thing to create to tell this story, while also really psyched to get to break everything down so clearly. As it’s come together, every codon in the 2D overlay has had to have it’s own layer in After Effects, but now that I finally have everything placed and animated, I’m really excited about it. Back in school, I recall one of our teachers, John Daugherty, telling us about how every piece that you do has to have that thing that makes it really interesting to you as an artist. Whether it’s a concept, or a technique, or whatever, and I think this part of this animation is that for me. It’s something that I don’t see other animations doing, and for me it’s like wrapping up that little aha moment that you get when you’re watching these sorts of things and you finally figure out how one representation relates to another and start to get what it is that you’re seeing.
As you can see, though, I’ve got these huge blocks of layers in the timeline there. At one point, I had more than 60 in there, and that’s with many of them being pre-compositions that lead to more layers in another timeline. But it’s working. And I just love it when things work.
This is going to be a good one.
Getting to Know Materials at the Molecular Level and Loving It
I made silicone! Ok, I made virtual silicone, but it’s still exciting to me. It was a work thing, so I hesitate to go into the details of the particular need, but the exciting part is that Marvin Sketch really is as cool as I thought it was, and I was able to draw out the individual atoms of a silicone polymer chain and produce a 3D model of that chain. And after working for years with varying kinds of silicones and studying their properties, that was just really cool for me. I’ve never really known the chemistry side of all of these materials I’ve been working with. I mean, I have notes somewhere about long polymer chains producing a different effect than the short ones (without looking it up, I want to say that the short ones were more brittle), but I really think that I could understand that so much better now with all the exposure to chemistry I am getting.
Oh drat, I wanted to show an image of the simple illustration you get out of Marvin Sketch, but it seems that the recent security hazards in allowing Java on a Mac (maybe this hits PCs too, I’m not sure), have my Java access shut down here at home for the time being. Anyway, I remain enthusiastic about this tool. And I’ll have it working from home again soon. And here, I’ll grab an image from Custom Silicone Rubberparts to give you an idea of what I mean. Hopefully they won’t mind because I am linking to them.
But yeah, we get to create models like that, which make me think about what it would take to literally make materials like that. It all starts with the knowledge, right?
And in the meantime I shake my fist at all those hackers and spammers out there that keep mucking up perfectly good tools for the rest of us. Java is a good tool. There’s no reason to mess with that. And all the spam comments I get here and even worse on my photo site, not cool. But making silicone, virtually, or for really really real, very cool. And all you real people who sometimes leave comments here, absolutely cool. 🙂
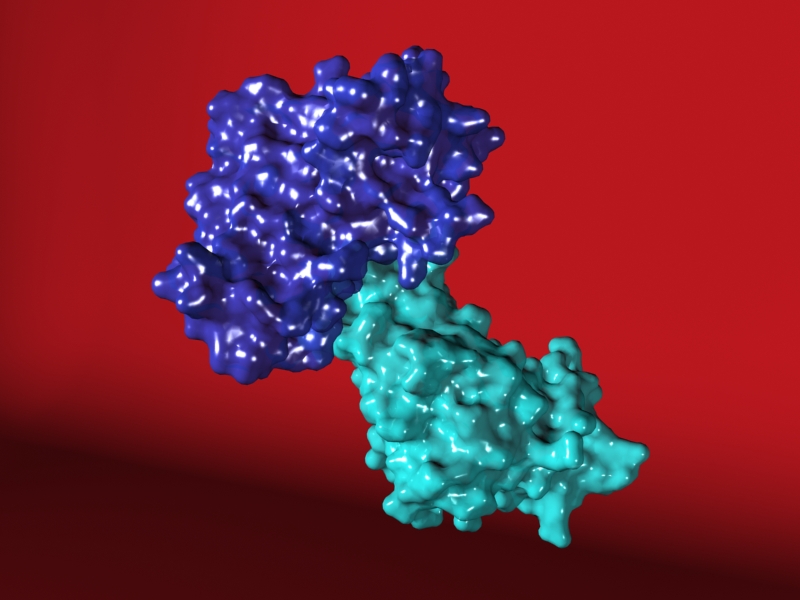
The Enzyme Substrate Complex
So, some of you may remember my recent post, “Molecules Molecules Molecules.” Well I’m really psyched to be able to share the animation that inspired that post with you all online now.
Enzyme Substrate Complex from sara egner on Vimeo.
I’m excited to get to show a little bit of the kind of work I’ve been doing lately over at Sapling. And I’m glad I get to share this one in particular because I know it goes just that extra step beyond the kinds of videos you find on this subject out there. And I’m loving that we get to make things like that. I get to work with biology experts. And when things get really really tiny, no one says, well that’s really just chemistry at that point – don’t worry about it. We just talk to a chemist about it. And if I hit a technological glitch, we have those kinds of experts around too. And I’d thought that my move into grad school was going to be a move away from all the video work I used to do. But here I am drawing back on all of that experience to make things like this now. And of course my whole little fascination with ATP comes in to play with this one. But that would be a whole other story. And this writer is winding down for the night.
Molecules Molecules Molecules!
This week, I’ve been engrossed in making molecular movie magic! When I was a student at UIC, we learned about the Protein Data Bank, and I thought that was pretty cool. We would bring files into a free program called Chimera, clean them up a bit, and export them as .stl or .obj files, anything we could pull into 3DsMax. I think that with larger files, you would also have to take the intermediate step of going into Mesh Lab to try and reduce your polygon count. From there, you could put any kind of material on them, set up lighting, do anything you wanted to.
I think that this was the first one I ever tried…
The image itself may not be all that impressive on it’s own right, but I remember being excited because I’d found something related to a good friend’s graduate work. I wish I could remember the name of it now, but I know it was tied to the magnetic bacteria that Cody studied when he was at CalTech. And just the thought of being able to search for and find 3D data related to a friend’s work like that, to be able to bring it into a 3D space and look at it, and then further still, be able to make something of it, and render it into an image that could be shared, well that all seemed pretty exciting.
A year or two later, they came out with Molecular Maya. This took out all the difficulty of working with Chimera, and allowed you to pull data directly from the PDB (Protein Data Bank) into Maya (which is another 3D animation program, much like 3DsMax).
Since coming to work for Sapling Learning, I have been using Cinema 4D (as opposed to either Maya or 3DsMax). And I have a plug-in downloaded called ePMV which is short for Embedded Python Molecule Viewer. It’s pretty awesome. Much like with Molecular Maya, you get to bring files directly in to your 3D scenes, without having to use other software in the interim.
I’m not sure if it didn’t exist before, or if I just didn’t know about it my first times trying to navigate the PDB, but finding PDB 101 was been a great help in being able to find the right proteins when looking for something specific. It’s also been a wealth of information when I’m trying to understand more about a molecule that I’m working with.
But I have to say that what I’m really excited about this week, is the realization that Marvin Sketch can be used to get a .pdb file on any molecule that you can draw. A coworker/chemistry genius showed me the Marvin Sketch Molecule Viewer, and I knew it was awesome when he did, but the more I get my head around all this, the more awesome this tool becomes. You can literally draw out any molecule you know the arrangement for, ask it to clean it up in either 2D or 3D for you, and if you’re looking at the 3D, you can export the file as a .pdb and just like that you have everything you need to bring it in via ePMV and use it for static illustration or animation.
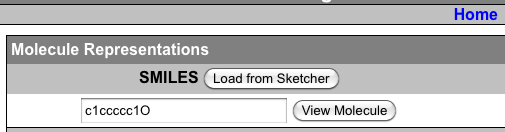
Now this still leaves me limited by the fact that I really don’t have a background in chemistry, and I often don’t know how to draw out the thing that I want (though I am learning a lot of that sort of stuff these days). So how amazing did I feel when just this week I noticed SMILES (Simplified molecular-input line-entry system.) Oh yeah. So there I am, scouring the internet for a clean model of ATP (adenosine triphosphate). And there are literally thousands of references to it within the PDB but it’s always with something. I wanted to find it by itself. And then I’m looking at the Wikipedia page on it. They have 3D models featured. Clearly this data is out there. I should be able to find it. And then I’m looking at the formula for ATP, and it’s C10H16N5O13P3. So I think to myself, that if I were better at drawing out molecules, I could just get it out of Marvin sketch. But sometimes Wikipedia has links to PDB pages, where was that? Well it didn’t have that but it did have a listing where you could show SMILES data
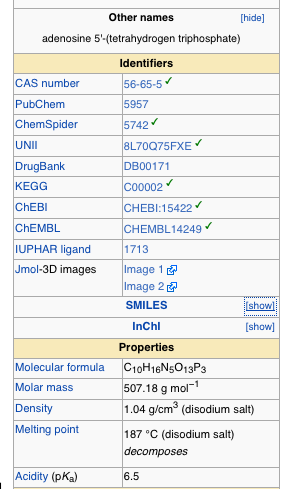 And if you just click the box that says show, next to SMILES, it gives you a code. You copy that code and paste it right into Marvin Sketch Molecule View SMILES loader
And if you just click the box that says show, next to SMILES, it gives you a code. You copy that code and paste it right into Marvin Sketch Molecule View SMILES loader
And just like that, you can get any molecule that you have that code for. I did this for ATP, ADP, glucose, and glucose 6 phosphate. They all had a SMILES code, listed right there in Wikipedia. And I was able to save .pdb files right my local computer’s desktop. And when I pulled them all in to Cinema 4D with the ePMV plug-in, I realized that it was easy to see exactly what atoms are coming off of one molecule and added to the other in the process I am working with. And because it was all so accessible like that, we’re going to be able to show people that, really clearly.
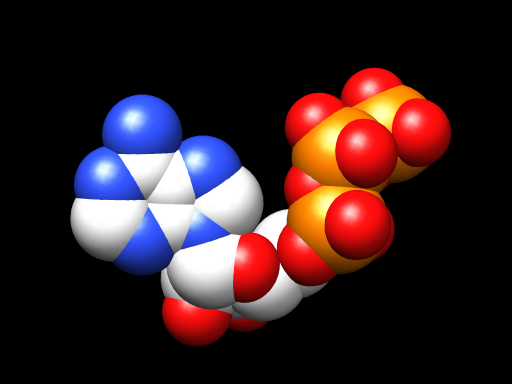
I don’t have any of my files from work handy here, but just now I put ATP’s code into the SMILES loader of Marvin Sketch and exported a 3D PDB file. I opened the file using Chimera and in less than a minute had this.
And if I were doing this in a 3D animation program, I could take things further still and make it pretty, make it move. Or I could grab that last bundle of spheres there, the last yellow (phosphorous) and the reds (oxygen atoms) clinging to it, and pop that right off to show you the difference between ATP and ADP, and what exactly that means when people talk about that happening.
I think that’s really friggin cool!
Well, I suppose I should stipulate that we’re *almost* seeing exactly how that works. I just learned today that there’s hydrogen in there too, but chemists seem to have this game of hide-the-hydrogen that they like to play on the rest of us. They claim it’s clearer that way. They’re probably right, but I’m not 100% sold yet. Still, except for the invisible hydrogen atoms, this is what ATP looks like. This is it’s shape. And all those reactions in the body where ATP gets converted to ADP are just this little guy getting a little bit shorter when that last phosphate group teams up somewhere else.
When I made my sliding filament animation, I remember having no idea what ATP actually looked like. I remember talking to other people about it, and none of the people I spoke to knew either. I wound up representing it with big yellow spheres in the animation. And that worked for the piece that it was, and was actually very clear in terms of what needed to be shown there. But I just love that this week, while trying to figure something out about ATP I wound up with not only a better picture of it, but my own model to play with and animate.
I suppose that lately it’s just been molecules molecules molecules for me. I never thought I would get this into the micro world, but I have to say, I think it was time. And with all this technology, all these programs making it easier than ever to really see and get a handle on what everything really is, well, I can’t think of a better time to be learning this stuff.